Experiment 2
Nonlinear effect of prediction error on recognition-based episodic memory updating
Taehoon Kim, Do-Joon Yi
2022-02-22
set.seed(12345)
if (!require("magick", quietly = TRUE)) install.packages("magick")
pacman::p_load(tidyverse, knitr,
afex, emmeans,
psych, ggplot2, papaja, cowplot)
pacman::p_load_gh("thomasp85/patchwork", "RLesur/klippy")
options(dplyr.summarise.inform=FALSE)
options(knitr.kable.NA = '')
set_sum_contrasts() # see Singmann & Kellen (2020)
klippy::klippy()1 Design & Procedure
f1 <- ggdraw() + draw_image("fig/F1A.png", scale = .9)
f2 <- ggdraw() + draw_image("fig/F5B.png", scale = .9)
plot_grid(f1, f2, labels = c('A', 'B'), nrow = 2, label_size = 20)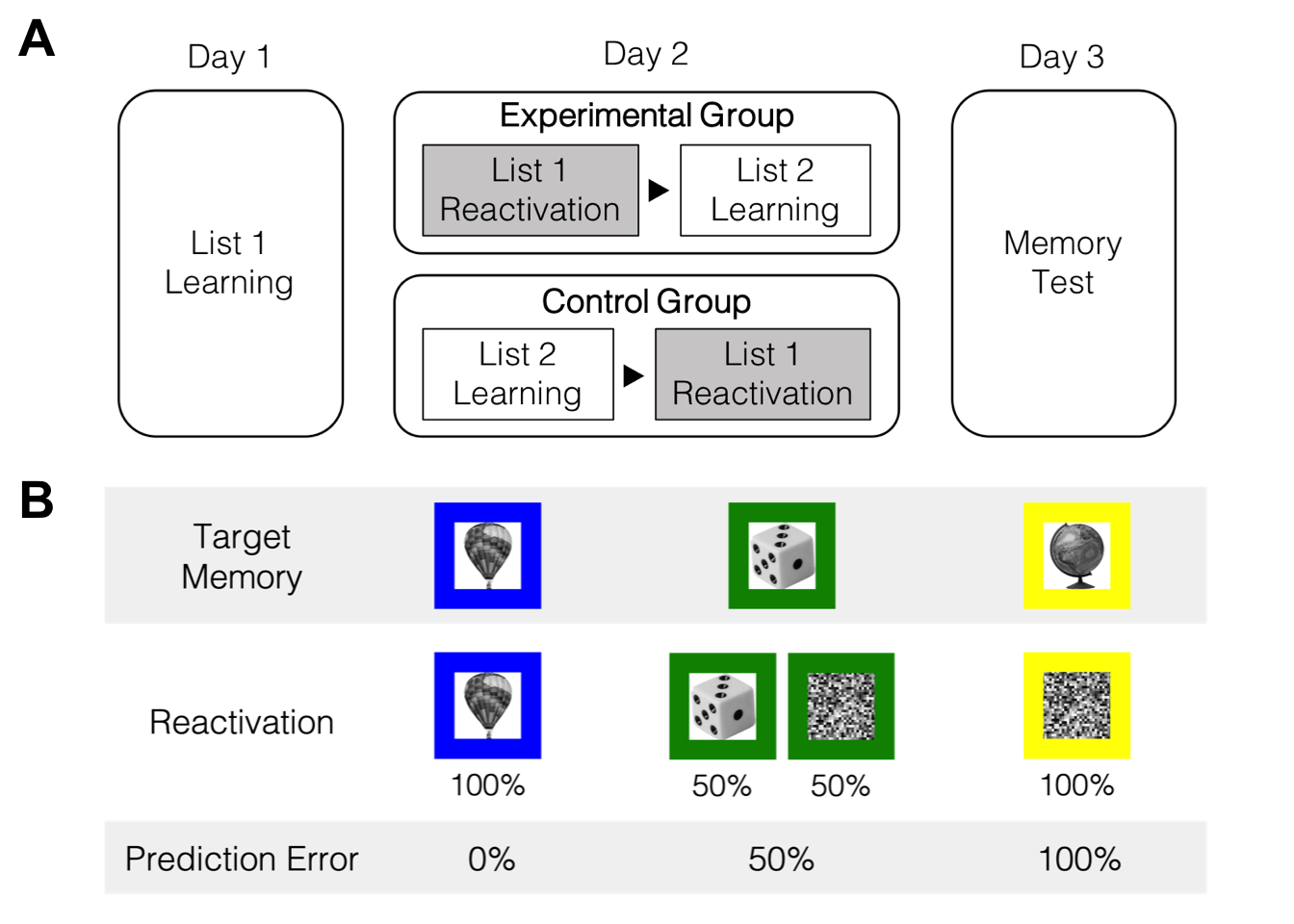
2 Day 1 & 2
첫째 날과 둘째 날에 학습한 List 1과 List 2의 물체-색상 연합학습 강도를 확인할 수 있다.
# Data
h1 <- read.csv("data/MemUdt_PE_d1t2_m.csv", header = T)
h2 <- read.csv("data/MemUdt_PE_d2t5_m.csv", header = T)
h1$List <- 'list1';
h2$List <- 'list2';
e1 <- rbind(h1, h2)
e1$SN <- factor(e1$SN)
e1$Group = factor(e1$Group, levels = c(1,2), labels=c("Experimental", "Control"))
e1$List <- factor(e1$List)
headTail(e1)
## SN Group Trial Block bTrial cCue IMidx IMname CueName
## 1 1 Experimental 1 1 1 1 147 man245.jpg cue_1.jpg
## 2 1 Experimental 2 1 2 1 56 man092.jpg cue_1.jpg
## 3 1 Experimental 3 1 3 1 79 man123.jpg cue_1.jpg
## 4 1 Experimental 4 1 4 2 100 man156.jpg cue_2.jpg
## ... <NA> <NA> ... ... ... ... ... <NA> <NA>
## 25917 38 Control 357 4 87 1 252 man439.jpg cue_1.jpg
## 25918 38 Control 358 4 88 1 129 man202.jpg cue_1.jpg
## 25919 38 Control 359 4 89 2 87 man133.jpg cue_2.jpg
## 25920 38 Control 360 4 90 3 19 man039.jpg cue_3.jpg
## Resp RT Corr List
## 1 1 0.84 1 list1
## 2 2 1.54 0 list1
## 3 1 1.6 1 list1
## 4 2 1.3 1 list1
## ... ... ... ... <NA>
## 25917 1 0.36 1 list2
## 25918 1 0.19 1 list2
## 25919 2 0.24 1 list2
## 25920 3 0.19 1 list2
table(e1$Group, e1$SN)
##
## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16
## Experimental 720 0 720 0 720 0 720 720 720 0 720 0 0 0 720 0
## Control 0 720 0 720 0 720 0 0 0 720 0 720 720 720 0 720
##
## 17 18 20 21 22 23 24 25 26 27 29 30 31 32 33 34
## Experimental 720 0 0 720 0 720 0 720 0 720 720 0 720 0 720 0
## Control 0 720 720 0 720 0 720 0 720 0 0 720 0 720 0 720
##
## 35 36 37 38
## Experimental 720 0 720 0
## Control 0 720 0 720
# descriptive
e1 %>% group_by(Group, SN, List, Block) %>%
summarise(Accuracy = mean(Corr)*100) %>%
ungroup() %>%
group_by(Group, List, Block) %>%
summarise(Accuracy = mean(Accuracy)) %>%
ungroup() %>%
pivot_wider(names_from = 'Block', values_from = 'Accuracy') %>%
kable(digits = 4, caption = "Descriptive statistics: Group x List x Black")| Group | List | 1 | 2 | 3 | 4 |
|---|---|---|---|---|---|
| Experimental | list1 | 65.8642 | 85.7407 | 93.4568 | 98.0247 |
| Experimental | list2 | 74.8765 | 91.2346 | 96.2346 | 98.6420 |
| Control | list1 | 59.5679 | 82.5926 | 92.2840 | 97.3457 |
| Control | list2 | 67.5926 | 87.4074 | 94.9383 | 97.2222 |
e1 %>% group_by(Group, SN, List, Block) %>%
summarise(Accuracy = mean(Corr)*100) %>%
ungroup() %>%
group_by(List, Block) %>%
summarise(Accuracy = mean(Accuracy)) %>%
ungroup() %>%
pivot_wider(names_from = 'Block', values_from = 'Accuracy') %>%
kable(digits = 4, caption = "Descriptive statistics: List x Black")| List | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| list1 | 62.7160 | 84.1667 | 92.8704 | 97.6852 |
| list2 | 71.2346 | 89.3210 | 95.5864 | 97.9321 |
# 3way ANOVA
e1.aov <- e1 %>% group_by(Group, SN, List, Block) %>%
summarise(Accuracy = mean(Corr)*100) %>%
ungroup() %>%
aov_ez(id = 'SN', dv = 'Accuracy',
between = 'Group', within = c('List', 'Block'),
anova_table = list(es = 'pes'))
e1.aov## Anova Table (Type 3 tests)
##
## Response: Accuracy
## Effect df MSE F pes p.value
## 1 Group 1, 34 224.67 3.16 + .085 .084
## 2 List 1, 34 34.73 35.86 *** .513 <.001
## 3 Group:List 1, 34 34.73 0.21 .006 .652
## 4 Block 1.96, 66.56 72.24 289.63 *** .895 <.001
## 5 Group:Block 1.96, 66.56 72.24 2.73 + .074 .074
## 6 List:Block 2.17, 73.89 17.83 17.37 *** .338 <.001
## 7 Group:List:Block 2.17, 73.89 17.83 0.05 .001 .964
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1
##
## Sphericity correction method: GGList 1보다 List 2의 정확도가 항상 높았지만, 그 차이는 후반부로 갈수록 감소한다.
e1.emm <- e1.aov %>%
emmeans(pairwise ~ List | Block, type = "response") %>%
summary(by = NULL, adjust = "bonferroni")
e1.emm[[2]]## contrast Block estimate SE df t.ratio p.value
## list1 - list2 X1 -8.519 1.478 34 -5.765 <.0001
## list1 - list2 X2 -5.154 1.122 34 -4.592 0.0002
## list1 - list2 X3 -2.716 0.730 34 -3.719 0.0029
## list1 - list2 X4 -0.247 0.324 34 -0.762 1.0000
##
## Results are averaged over the levels of: Group
## P value adjustment: bonferroni method for 4 tests실험 1과 달리, List 1과 2의 차이는 마지막 구획에서 유의미하지 않다.
3 Day 3
# Data
e2 <- read.csv("data/MemUdt_PE_d3t6_m.csv", header = T)
e2$SN <- factor(e2$SN)
e2$Group <- factor(e2$Group, levels=c(1,2), labels=c("Experimental","Control"))
e2$List <- factor(e2$List, levels=c(1,2,3), labels=c("list1","list2","list3"))
e2$Cue <- factor(e2$Cue, levels=c(1,2,3,0), labels=c("cc1","cc2","cc3","lure"))
# e2$PE <- factor(e2$PE, levels=c(1,2,3,0), labels=c("pe0","pe50","pe100","lure"))
e2$CueName = factor(e2$CueName, labels=c("c0","c1","c2","c3"))
e2$Resp <- factor(e2$Resp, levels=c(1,2,3), labels=c("list1", "list2", "list3"))
e2$PE[e2$PE==0] <- NA
e2$PE[e2$PE==1] <- 0
e2$PE[e2$PE==2] <- 50
e2$PE[e2$PE==3] <- 100
str(e2)
## 'data.frame': 9720 obs. of 19 variables:
## $ SN : Factor w/ 36 levels "1","2","3","4",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Group : Factor w/ 2 levels "Experimental",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ Trial : int 1 2 3 4 5 6 7 8 9 10 ...
## $ List : Factor w/ 3 levels "list1","list2",..: 3 1 1 2 1 3 1 1 1 2 ...
## $ Cue : Factor w/ 4 levels "cc1","cc2","cc3",..: 4 2 2 2 2 4 3 3 2 3 ...
## $ PE : num NA 50 50 50 50 NA 0 0 50 0 ...
## $ IMidx : int 72 38 87 95 129 240 65 71 253 201 ...
## $ IMname : chr " man114.jpg" " man066.jpg" " man133.jpg" " man145.jpg" ...
## $ CueName: Factor w/ 4 levels "c0","c1","c2",..: 1 3 3 3 3 1 4 4 3 4 ...
## $ Resp : Factor w/ 3 levels "list1","list2",..: 3 1 1 2 1 3 2 2 2 2 ...
## $ RT : num 2.81 3.62 5.18 1.61 1.42 ...
## $ Corr : int 1 1 1 1 1 1 2 2 2 1 ...
## $ Conf : int 4 4 4 4 4 4 4 4 4 3 ...
## $ cRT : num 2.487 1.26 0.876 0.981 0.891 ...
## $ aResp : int 7 2 2 2 2 7 3 1 2 3 ...
## $ aRT : num 0 4.19 0.671 0.927 0.544 ...
## $ aCorr : int 7 1 1 1 1 7 1 0 1 1 ...
## $ aConf : int 7 4 4 4 4 7 4 4 4 4 ...
## $ aCRT : num 0 1.604 1.74 0.803 0.463 ...마지막 날(Day 3)에 실시한 출처기억 검사 결과를 정리하였다.
3.1 Descriptive Stats
3.1.1 List 1 & List 2
# recognition: except List 3
e2old <- e2 %>%
filter(List == "list1" | List == "list2") %>%
select(SN, Group, List, Cue, PE, Resp, Corr, Conf) %>%
droplevels()
unique(e2old$List)
## [1] list1 list2
## Levels: list1 list2
unique(e2old$Resp)
## [1] list1 list2 list3
## Levels: list1 list2 list3
unique(e2old$Corr)
## [1] 1 2 3 0
e2old$Miss <- as.numeric(e2old$Corr==0) # recognition: miss rate
e2old$Correct <- as.numeric(e2old$Corr==1) # correct source memory
e2old$L1toL2 <- as.numeric(e2old$Corr==2) # source confusion
e2old$L2toL1 <- as.numeric(e2old$Corr==3) # source confusion + intrusion
glimpse(e2old)
## Rows: 6,480
## Columns: 12
## $ SN <fct> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
## $ Group <fct> Experimental, Experimental, Experimental, Experimental, Experi…
## $ List <fct> list1, list1, list2, list1, list1, list1, list1, list2, list2,…
## $ Cue <fct> cc2, cc2, cc2, cc2, cc3, cc3, cc2, cc3, cc2, cc2, cc1, cc1, cc…
## $ PE <dbl> 50, 50, 50, 50, 0, 0, 50, 0, 50, 50, 100, 100, 100, 100, 100, …
## $ Resp <fct> list1, list1, list2, list1, list2, list2, list2, list2, list2,…
## $ Corr <int> 1, 1, 1, 1, 2, 2, 2, 1, 1, 1, 1, 1, 3, 3, 1, 1, 1, 1, 3, 3, 1,…
## $ Conf <int> 4, 4, 4, 4, 4, 4, 4, 3, 4, 4, 4, 3, 4, 3, 4, 4, 4, 3, 3, 3, 4,…
## $ Miss <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ Correct <dbl> 1, 1, 1, 1, 0, 0, 0, 1, 1, 1, 1, 1, 0, 0, 1, 1, 1, 1, 0, 0, 1,…
## $ L1toL2 <dbl> 0, 0, 0, 0, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
## $ L2toL1 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 1, 1, 0,…
e2oldslong <- e2old %>%
group_by(Group, SN, List) %>%
summarise(Correct = mean(Correct)*100,
Miss = mean(Miss)*100,
L1toL2 = mean(L1toL2)*100,
L2toL1 = mean(L2toL1)*100) %>%
ungroup() %>%
mutate(AttrError = L1toL2 + L2toL1) %>%
select(Group, SN, List, Correct, Miss, AttrError)
e2oldslong %>% group_by(Group, List) %>%
summarise(Correct = mean(Correct),
Miss = mean(Miss),
AttrError = mean(AttrError)) %>%
ungroup() %>%
kable(digits = 4, caption = "Descriptive statistics: Group x List")| Group | List | Correct | Miss | AttrError |
|---|---|---|---|---|
| Experimental | list1 | 75.4321 | 3.2716 | 21.2963 |
| Experimental | list2 | 66.0494 | 2.2222 | 31.7284 |
| Control | list1 | 71.4198 | 4.6296 | 23.9506 |
| Control | list2 | 70.6790 | 3.5802 | 25.7407 |
3.1.2 List 3
e2new <- e2 %>%
filter(List == "list3") %>%
select(SN, Group, Resp, Corr, Conf) %>%
droplevels()
head(e2new)
## SN Group Resp Corr Conf
## 1 1 Experimental list3 1 4
## 2 1 Experimental list3 1 4
## 3 1 Experimental list3 1 4
## 4 1 Experimental list3 1 4
## 5 1 Experimental list3 1 4
## 6 1 Experimental list3 1 2
unique(e2new$Resp)
## [1] list3 list1 list2
## Levels: list1 list2 list3
unique(e2new$Corr)
## [1] 1 0
e2new %>% group_by(Group, SN) %>%
summarise(FA = 100 - mean(Corr)*100) %>%
ungroup() %>%
group_by(Group) %>%
summarise(FA = mean(FA)) %>%
ungroup() %>%
kable(digits = 4, caption = "Descriptive statistics: Group")| Group | FA |
|---|---|
| Experimental | 2.7160 |
| Control | 4.3827 |
3.2 Miss
e2oldslong %>% aov_ez(id = 'SN', dv = 'Miss',
between = 'Group', within = 'List',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: Miss
## Effect df MSE F pes p.value
## 1 Group 1, 34 13.13 2.53 .069 .121
## 2 List 1, 34 6.93 2.86 .078 .100
## 3 Group:List 1, 34 6.93 0.00 <.001 >.999
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1List 1과 List 2의 항목을 “본 적 없다”고 답하는 비율은 매우 낮았고, 조건간 차이도 유의미하지 않았다.
3.3 False alarm
e2new %>% group_by(Group, SN) %>%
summarise(FA = 100 - mean(Corr)*100) %>%
ungroup() %>%
aov_ez(id = 'SN', dv = 'FA', between = 'Group',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: FA
## Effect df MSE F pes p.value
## 1 Group 1, 34 30.26 0.83 .024 .370
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1List 3의 항목을 “봤다”고 답한 비율도 낮았다. 집단 차이는 유의미하지 않았다.
3.4 Source accuracy
e2oldslong %>% aov_ez(id = 'SN', dv = 'Correct',
between = 'Group', within = 'List',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: Correct
## Effect df MSE F pes p.value
## 1 Group 1, 34 118.77 0.01 <.001 .905
## 2 List 1, 34 109.56 4.21 * .110 .048
## 3 Group:List 1, 34 109.56 3.07 + .083 .089
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 13.4.0.1 Source misattribution
e2AttrErr.aov <- e2oldslong %>%
aov_ez(id = 'SN', dv = 'AttrError',
between = 'Group', within = 'List',
anova_table = list(es = 'pes'))
e2AttrErr.aov ## Anova Table (Type 3 tests)
##
## Response: AttrError
## Effect df MSE F pes p.value
## 1 Group 1, 34 81.07 0.62 .018 .438
## 2 List 1, 34 102.44 6.56 * .162 .015
## 3 Group:List 1, 34 102.44 3.28 + .088 .079
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1# custom contrast
e2AttrErr.emm <- e2AttrErr.aov %>% emmeans(~ List*Group)
con <- emmeans:::trt.vs.ctrl.emmc(1:4)
ExpL2L1 <- c(-1, 1,0,0)
ConL2L1 <- c(0,0,-1, 1)
L2ExpCon <- c(0,1,0,-1)
con <- data.frame(ExpL2L1, ConL2L1, L2ExpCon)
contrast(e2AttrErr.emm, con, adjust = "bonferroni")## contrast estimate SE df t.ratio p.value
## ExpL2L1 10.43 3.37 34 3.092 0.0119
## ConL2L1 1.79 3.37 34 0.531 1.0000
## L2ExpCon 5.99 3.49 34 1.716 0.2860
##
## P value adjustment: bonferroni method for 3 testsANOVA에서 List 2를 List 1으로 오기억하는 경우가 반대보다 높았으나, 집단 차이는 유의미하지 않았다. 그러나 사후비교에서 실험집단의 경우에만 List 2 오귀인이 List 1 오귀인보다 높았다(비대칭적 오귀인 분포).
3.5 Prediction error effects
e2PEslong <- e2old %>%
filter(List == 'list2') %>%
group_by(Group, SN, PE) %>%
droplevels() %>%
summarise(L2toL1 = mean(L2toL1)) %>%
ungroup()
str(e2PEslong)
## tibble [108 × 4] (S3: tbl_df/tbl/data.frame)
## $ Group : Factor w/ 2 levels "Experimental",..: 1 1 1 1 1 1 1 1 1 1 ...
## $ SN : Factor w/ 36 levels "1","2","3","4",..: 1 1 1 3 3 3 5 5 5 7 ...
## $ PE : num [1:108] 0 50 100 0 50 100 0 50 100 0 ...
## $ L2toL1: num [1:108] 0.2 0.3 0.533 0.167 0.233 ...e2PEslong %>%
group_by(Group, PE) %>%
summarise(L2toL1 = mean(L2toL1)) %>%
ungroup() %>%
pivot_wider(names_from = 'PE', values_from = 'L2toL1') %>%
kable(digits = 4, caption = "Descriptive statistics: Group x PE")| Group | 0 | 50 | 100 |
|---|---|---|---|
| Experimental | 0.2667 | 0.3648 | 0.3204 |
| Control | 0.2519 | 0.2426 | 0.2778 |
e2PE.aov <- e2PEslong %>%
aov_ez(id = 'SN', dv = 'L2toL1',
between = 'Group', within = 'PE',
anova_table = list(es = 'pes'))
e2PE.aov## Anova Table (Type 3 tests)
##
## Response: L2toL1
## Effect df MSE F pes p.value
## 1 Group 1, 34 0.03 2.94 + .080 .095
## 2 PE 1.98, 67.46 0.01 2.87 + .078 .064
## 3 Group:PE 1.98, 67.46 0.01 3.73 * .099 .029
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1
##
## Sphericity correction method: GG집단과 예측오류 수준의 상호작용이 유의미하였다.
# custom contrast
e2PE.emm <- e2PE.aov %>% emmeans(~ Group*PE)
Exp0to50 <- c(-1,0,1,0,0,0)
Exp0to100 <- c(-1,0,0,0,1,0)
Exp100to50 <- c(0,0,1,0,-1,0)
Con0to50 <- c(0,-1,0,1,0,0)
Con0to100 <- c(0,-1,0,0,0,1)
Con100to50 <- c(0,0,0,1,0,-1)
ExpCon50 <- c(0,0,1,-1,0,0)
con <- data.frame(Exp0to50, Exp0to100, Exp100to50,
Con0to50, Con0to100, Con100to50, ExpCon50)
contrast(e2PE.emm, con, adjust = "Bonferroni")## contrast estimate SE df t.ratio p.value
## Exp0to50 0.09815 0.0287 34 3.424 0.0114
## Exp0to100 0.05370 0.0301 34 1.787 0.5799
## Exp100to50 0.04444 0.0278 34 1.596 0.8377
## Con0to50 -0.00926 0.0287 34 -0.323 1.0000
## Con0to100 0.02593 0.0301 34 0.863 1.0000
## Con100to50 -0.03519 0.0278 34 -1.264 1.0000
## ExpCon50 0.12222 0.0412 34 2.969 0.0381
##
## P value adjustment: bonferroni method for 7 tests실험집단에서만 PE50의 침범반응이 PE0보다 컸다.
emm.t2 <- e2PE.aov %>% emmeans(pairwise ~ PE | Group)
emm.t2[[1]] %>% contrast("poly") %>%
summary(by = NULL, adjust = "Bonferroni")## contrast Group estimate SE df t.ratio p.value
## linear Experimental 0.0537 0.0301 34 1.787 0.3313
## quadratic Experimental -0.1426 0.0479 34 -2.979 0.0212
## linear Control 0.0259 0.0301 34 0.863 1.0000
## quadratic Control 0.0444 0.0479 34 0.929 1.0000
##
## P value adjustment: bonferroni method for 4 testsemm.t2[[1]] %>% contrast(interaction = c("poly", "consec"),
by = NULL, adjust = "Bonferroni")## PE_poly Group_consec estimate SE df t.ratio p.value
## linear Control - Experimental -0.0278 0.0425 34 -0.654 1.0000
## quadratic Control - Experimental 0.1870 0.0677 34 2.763 0.0183
##
## P value adjustment: bonferroni method for 2 tests이차함수 추세(quadratic trend)에서 PE와 집단의 상호작용이 유의미하였다(가정 중요한 결과).
3.6 Source confidence
e2old %>% filter(Corr == 2 | Corr == 3) %>%
droplevels() %>%
group_by(Group, SN, List) %>%
summarise(Confid = mean(Conf)) %>%
ungroup() %>%
group_by(Group, List) %>%
summarise(Confid = mean(Confid)) %>%
ungroup() %>%
pivot_wider(names_from = 'List', values_from = 'Confid') %>%
kable(digits = 4, caption = "Descriptive statistics: Group x List")| Group | list1 | list2 |
|---|---|---|
| Experimental | 2.7256 | 3.0859 |
| Control | 2.7576 | 3.0635 |
e2old %>% filter(Corr == 2 | Corr == 3) %>%
droplevels() %>%
group_by(Group, SN, List) %>%
summarise(Confid = mean(Conf)) %>%
ungroup() %>%
aov_ez(id = 'SN', dv = 'Confid',
between = 'Group', within = 'List',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: Confid
## Effect df MSE F pes p.value
## 1 Group 1, 34 0.42 0.00 <.001 .975
## 2 List 1, 34 0.06 32.43 *** .488 <.001
## 3 Group:List 1, 34 0.06 0.22 .006 .644
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1List 1보다 List 2에 대한 확신도가 더 높았다.
# List 2 misattr X PE: Confidence
e2old %>% filter(List == 'list2') %>%
droplevels() %>%
group_by(Group, SN, PE) %>%
summarise(Conf = mean(Conf)) %>%
ungroup() %>%
group_by(Group, PE) %>%
summarise(Conf = mean(Conf)) %>%
ungroup() %>%
pivot_wider(names_from = 'PE', values_from = 'Conf') %>%
kable(digits = 4, caption = "Descriptive statistics: Group x PE")| Group | 0 | 50 | 100 |
|---|---|---|---|
| Experimental | 3.1222 | 3.1981 | 3.1630 |
| Control | 3.1426 | 3.1667 | 3.1278 |
e2old %>% filter(List == 'list2') %>%
droplevels() %>%
group_by(Group, SN, PE) %>%
summarise(Conf = mean(Conf)) %>%
ungroup() %>%
aov_ez(id = 'SN', dv = 'Conf',
between = 'Group', within = 'PE',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: Conf
## Effect df MSE F pes p.value
## 1 Group 1, 34 0.61 0.01 <.001 .919
## 2 PE 1.93, 65.62 0.03 0.83 .024 .435
## 3 Group:PE 1.93, 65.62 0.03 0.30 .009 .735
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 1
##
## Sphericity correction method: GG3.7 Associative Memory
3.7.1 Accuracy
e2 %>% filter(List != "list3", Resp != "list3") %>%
group_by(Group, SN, List) %>%
summarise(Accuracy = mean(aCorr)*100) %>%
ungroup() %>%
group_by(Group, List) %>%
summarise(Accuracy = mean(Accuracy)) %>%
ungroup() %>%
pivot_wider(names_from = 'List', values_from = 'Accuracy') %>%
kable(digits = 4, caption = "Descriptive statistics: Group x List")| Group | list1 | list2 |
|---|---|---|
| Experimental | 86.1648 | 89.9063 |
| Control | 85.2086 | 87.4866 |
e2 %>% filter(List != "list3", Resp != "list3") %>%
group_by(Group, SN, List) %>%
summarise(Accuracy = mean(aCorr)*100) %>%
ungroup() %>%
aov_ez(id = 'SN', dv = 'Accuracy',
between = 'Group', within = 'List',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: Accuracy
## Effect df MSE F pes p.value
## 1 Group 1, 34 107.50 0.48 .014 .494
## 2 List 1, 34 18.78 8.68 ** .203 .006
## 3 Group:List 1, 34 18.78 0.51 .015 .479
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 13.7.2 Confidence
e2 %>% filter(List != "list3", Resp != "list3") %>%
group_by(Group, SN, List) %>%
summarise(Confident = mean(aConf)) %>%
ungroup() %>%
group_by(Group, List) %>%
summarise(Confident = mean(Confident)) %>%
ungroup() %>%
pivot_wider(names_from = 'List', values_from = 'Confident') %>%
kable(digits = 4, caption = "Descriptive statistics: Group x List")| Group | list1 | list2 |
|---|---|---|
| Experimental | 3.5065 | 3.5713 |
| Control | 3.4765 | 3.5287 |
e2 %>% filter(List != "list3", Resp != "list3") %>%
group_by(Group, SN, List) %>%
summarise(Confident = mean(aConf)) %>%
ungroup() %>%
aov_ez(id = 'SN', dv = 'Confident',
between = 'Group', within = 'List',
anova_table = list(es = 'pes'))## Anova Table (Type 3 tests)
##
## Response: Confident
## Effect df MSE F pes p.value
## 1 Group 1, 34 0.24 0.10 .003 .753
## 2 List 1, 34 0.02 2.68 .073 .111
## 3 Group:List 1, 34 0.02 0.03 <.001 .861
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '+' 0.1 ' ' 14 Plots
e2cnt <- e2old %>%
group_by(Group, SN, List, Resp) %>%
summarise(n = n()) %>%
mutate(prop = n/sum(n)) %>%
ungroup() %>%
filter(Resp != 'list3') %>%
droplevels()
# print(e2cnt, n=20)
tmp0 <- e2cnt %>%
group_by(Group, List, Resp) %>%
summarise(MN = mean(prop),
SD = sd(prop)) %>%
ungroup()
tmp1 <- e2cnt %>%
filter(Group == 'Experimental') %>%
droplevels() %>%
papaja::wsci(id = 'SN',
factor = c('List', 'Resp'),
dv = 'prop') %>%
mutate(Group = "Experimental") %>%
rename("wsci" = "prop")
tmp2 <- e2cnt %>%
filter(Group == 'Control') %>%
droplevels() %>%
papaja::wsci(id = 'SN',
factor = c('List', 'Resp'),
dv = 'prop') %>%
mutate(Group = "Control") %>%
rename("wsci" = "prop")
tmp3 <- merge(tmp1, tmp2, all = TRUE)
e2cnt.g <- merge(tmp0, tmp3, by = c("Group", "List", "Resp"), all = TRUE)
F3A <- ggplot(data=e2cnt.g, aes(x=List, y=MN, fill=Resp)) +
geom_bar(stat='identity', width=0.7, color="black",
position=position_dodge(.8)) +
geom_errorbar(aes(x=List, ymin=MN-wsci, ymax=MN+wsci, group=Resp),
position = position_dodge(0.8), width=0.3,
show.legend = FALSE) +
facet_grid(.~Group) +
scale_x_discrete(labels=c("List 1","List 2")) +
scale_y_continuous(breaks=c(0, .2, .4, .6, .8, 1)) +
scale_fill_manual(values = c("#feb24c", "#91bfdb"),
labels = c("Day 1", "Day 2")) +
labs(x = "Object List", y = "Proportion", fill='Response') +
coord_cartesian(ylim = c(0, 1), clip = "on") +
theme_bw(base_size = 18) +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank())
F3A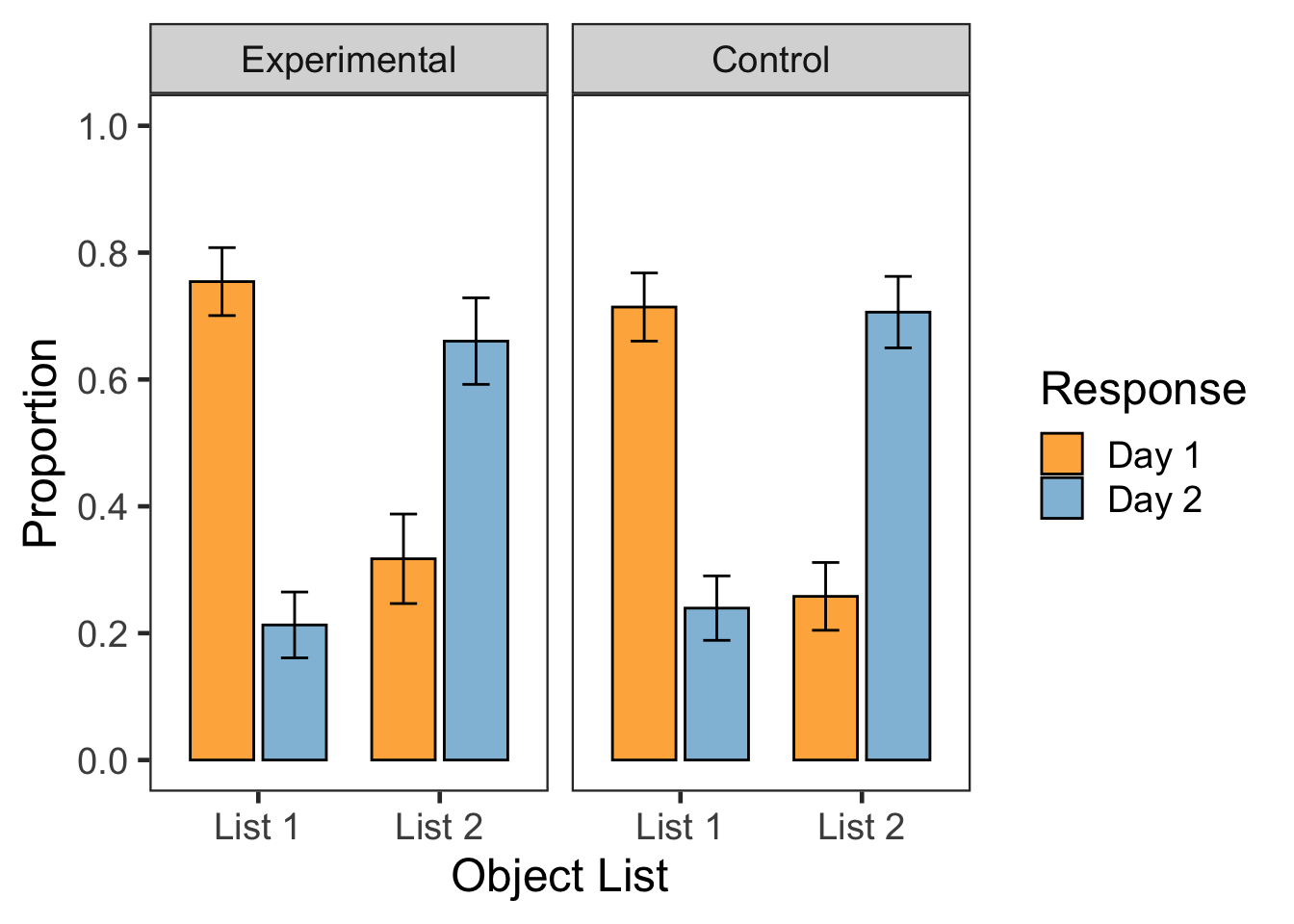
tmp0 <- e2PEslong %>%
group_by(Group, PE) %>%
summarise(MN = mean(L2toL1)) %>%
ungroup()
tmp1 <- e2PEslong %>%
filter(Group == 'Experimental') %>%
droplevels() %>%
papaja::wsci(id = 'SN',
factor = 'PE',
dv = 'L2toL1') %>%
mutate(Group = "Experimental") %>%
rename("wsci" = "L2toL1")
tmp2 <- e2PEslong %>%
filter(Group == 'Control') %>%
droplevels() %>%
papaja::wsci(id = 'SN',
factor = 'PE',
dv = 'L2toL1') %>%
mutate(Group = "Control") %>%
rename("wsci" = "L2toL1")
tmp3 <- merge(tmp1, tmp2, all = TRUE)
e2PEg <- merge(tmp0, tmp3, by = c("Group", "PE"), all = TRUE)
F3B <- ggplot(data=e2PEg, aes(x=PE, y=MN, group=Group,
ymin=MN-wsci, ymax=MN+wsci)) +
geom_line(position = position_dodge(width=10)) +
geom_errorbar(position = position_dodge(10), width=5,
show.legend = FALSE) +
geom_point(aes(shape=Group, fill=Group),
size=4, color='black', stroke=1,
position=position_dodge(width=10)) +
scale_x_continuous(breaks=c(0, 50, 100)) +
scale_y_continuous(breaks=c(0, .1, .2, .3, .4, .5)) +
scale_shape_manual(values = c(21, 23)) +
scale_fill_manual(values = c("black", "white")) +
labs(x = "Prediction Error (%)", y = "Proportion Intrusion", fill='Group') +
coord_cartesian(ylim = c(0, 0.5), clip = "on") +
theme_bw(base_size = 18) +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
aspect.ratio = 1)
F3B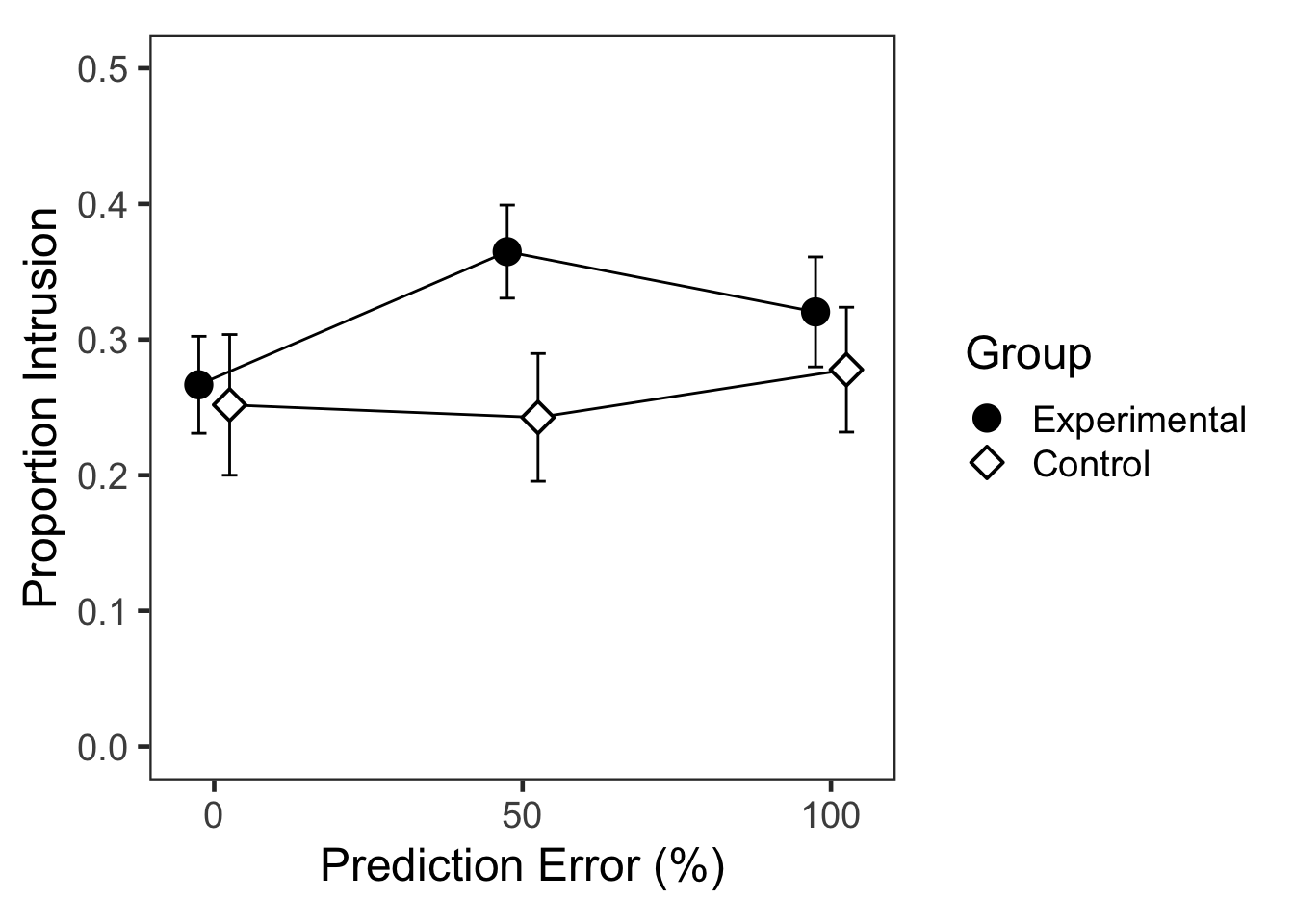
# all plots
# cowplot::plot_grid(F2A, F3A, ncol = 1, labels = c('A', 'B'), label_size = 20)
# cowplot::plot_grid(F2B, F3B, ncol = 1, labels = c('A', 'B'), label_size = 20)
# https://www.datanovia.com/en/blog/how-to-add-p-values-to-ggplot-facets/5 Session Info
sessionInfo()
## R version 4.1.2 (2021-11-01)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Monterey 12.2.1
##
## Matrix products: default
## LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] klippy_0.0.0.9500 patchwork_1.1.1 cowplot_1.1.1 papaja_0.1.0.9997
## [5] psych_2.1.9 emmeans_1.7.2 afex_1.0-1 lme4_1.1-28
## [9] Matrix_1.4-0 knitr_1.37 forcats_0.5.1 stringr_1.4.0
## [13] dplyr_1.0.8 purrr_0.3.4 readr_2.1.2 tidyr_1.2.0
## [17] tibble_3.1.6 ggplot2_3.3.5 tidyverse_1.3.1 magick_2.7.3
## [21] icons_0.2.0
##
## loaded via a namespace (and not attached):
## [1] TH.data_1.1-0 minqa_1.2.4 colorspace_2.0-3
## [4] ellipsis_0.3.2 rsconnect_0.8.25 rprojroot_2.0.2
## [7] estimability_1.3 fs_1.5.2 rstudioapi_0.13
## [10] farver_2.1.0 remotes_2.4.2 fansi_1.0.2
## [13] mvtnorm_1.1-3 lubridate_1.8.0 xml2_1.3.3
## [16] codetools_0.2-18 splines_4.1.2 mnormt_2.0.2
## [19] cachem_1.0.6 pkgload_1.2.4 jsonlite_1.7.3
## [22] nloptr_2.0.0 broom_0.7.12 dbplyr_2.1.1
## [25] compiler_4.1.2 httr_1.4.2 backports_1.4.1
## [28] assertthat_0.2.1 fastmap_1.1.0 cli_3.2.0
## [31] htmltools_0.5.2 prettyunits_1.1.1 tools_4.1.2
## [34] lmerTest_3.1-3 coda_0.19-4 gtable_0.3.0
## [37] glue_1.6.1 reshape2_1.4.4 rappdirs_0.3.3
## [40] Rcpp_1.0.8 carData_3.0-5 cellranger_1.1.0
## [43] jquerylib_0.1.4 vctrs_0.3.8 nlme_3.1-155
## [46] xfun_0.29 ps_1.6.0 brio_1.1.3
## [49] testthat_3.1.2 rvest_1.0.2 lifecycle_1.0.1
## [52] pacman_0.5.1 devtools_2.4.3 MASS_7.3-55
## [55] zoo_1.8-9 scales_1.1.1 hms_1.1.1
## [58] parallel_4.1.2 sandwich_3.0-1 yaml_2.3.5
## [61] curl_4.3.2 memoise_2.0.1 sass_0.4.0
## [64] stringi_1.7.6 highr_0.9 desc_1.4.0
## [67] boot_1.3-28 pkgbuild_1.3.1 rlang_1.0.1
## [70] pkgconfig_2.0.3 evaluate_0.15 lattice_0.20-45
## [73] labeling_0.4.2 processx_3.5.2 tidyselect_1.1.2
## [76] plyr_1.8.6 magrittr_2.0.2 R6_2.5.1
## [79] generics_0.1.2 multcomp_1.4-18 DBI_1.1.2
## [82] pillar_1.7.0 haven_2.4.3 withr_2.4.3
## [85] survival_3.2-13 abind_1.4-5 modelr_0.1.8
## [88] crayon_1.5.0 car_3.0-12 utf8_1.2.2
## [91] tmvnsim_1.0-2 tzdb_0.2.0 rmarkdown_2.11
## [94] usethis_2.1.5 grid_4.1.2 readxl_1.3.1
## [97] callr_3.7.0 reprex_2.0.1 digest_0.6.29
## [100] xtable_1.8-4 numDeriv_2016.8-1.1 munsell_0.5.0
## [103] bslib_0.3.1 sessioninfo_1.2.2Copyright © 2022 CogNIPS. All rights reserved.